Introduction
The use of animal models plays a crucial role in the advancement of biomedicine and has been a cornerstone of medical progress for many centuries. Mammals such as mice and monkeys have traditionally been the preferred models for biomedical research due to their close evolutionary relationship with humans. However, the use of mammals in research raises ethical and budgetary concerns and has led many laboratories and agencies to employ the principles first described by W. M. S. Russell and R. L. Burch as the “Three R’s”: Replacement, Reduction, and Refinement (Russell and Burch, 1959). These principles encourage the use of alternative model systems such as cell culture, Danio rerio (zebrafish), Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (worm) in the exploration of basic biology.
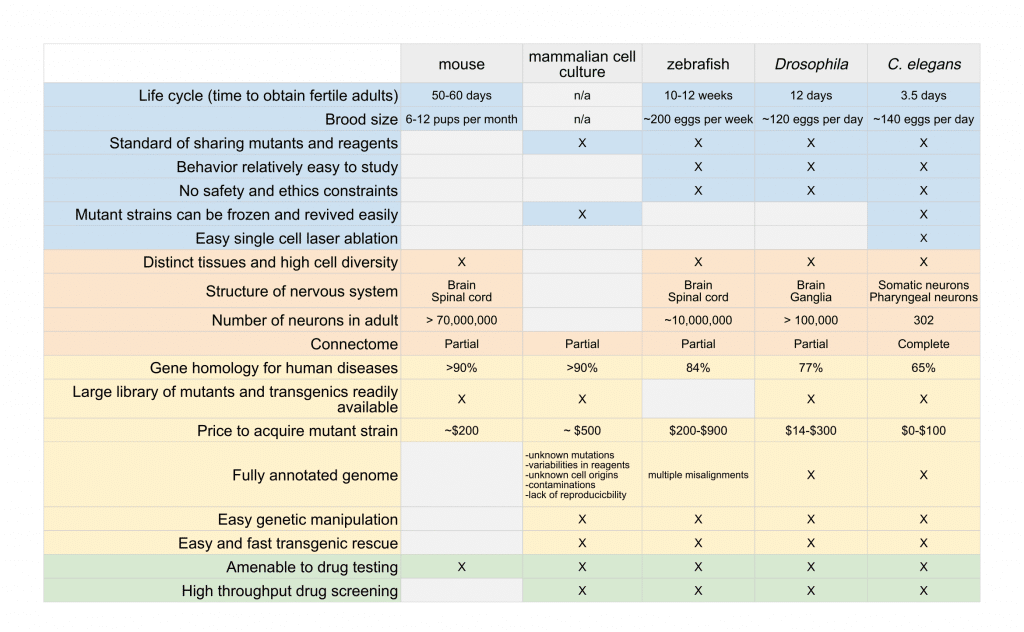
The selection of which organisms to use as a research model depends on the nature of the disease or mechanism being studied and the scientific questions being asked. In this article, we provide a comparison of the advantages and limitations of many commonly used model organisms (Table 1).
Small model systems in biomedical research
Cell cultures
Cell cultures are typically performed on isolated mammalian cells. They provide information about the interaction between molecules within a eukaryotic system. However, although such assays represent a fast and powerful tool for proof of concept, results obtained in vitro often poorly correlate with in vivo mechanisms. This is due in part to the fact the mechanisms studied in vitro occur in a simplified, highly controlled but often poorly understood intracellular and extracellular environment with little physiological relevance. In vivo, biological mechanisms and reactions depend both on the metabolism of the compounds and the interplay between organs within the whole animal. Moreover, the use of dissociated cells precludes access to the behavioral output of biological mechanisms.
C. elegans
C. elegans is a transparent, free-living, non-parasitic nematode. Both in biological and biomedical studies, C. elegans provides many advantages over vertebrates such as mouse and zebrafish models (Table 1). C. elegans hatched larvae are 0.25 millimeters long and adults are 1 millimeter long with a maximum diameter of ~80 µm, allowing the culture set-up to be minimal. This nematode can grow on agar medium in Petri dishes, where it can be maintained on a simple diet of Escherichia coli bacteria, which makes it very easy and cost-effective to grow in the laboratory. C. elegans has a short life cycle (3.5 days to obtain fertile adults at 20°C) and adult hermaphrodites are highly prolific reproducers (>140 eggs per adult and per day (Muschiol et al., 2009)). Males are largely absent in laboratory populations (1 out of 1000 individuals) and the hermaphrodites reproduce by self-fertilization, creating genetic clones of themselves.
C. elegans‘ architectural simplicity and lack of a central nervous system may be cited as a shortcoming when it comes to using the worm as a model to study mammalian genetic aberrations and drug efficiency. However, this simplicity has lead to one of the worm’s key advantages: all neurons have been identified and the connections between them are known. This neuronal map, referred to as the “connectome”, has not been completed for any other organism. In addition, with the use of in vivo fluorescent markers, processes such as axon growth, embryogenesis and fat metabolism can easily be studied in the living worm. Although the adult hermaphrodite has only 959 somatic cells and a nervous system containing 302 neuron, C. elegans is a sophisticated multicellular animal with many different organs and tissues including muscle, hypodermis (skin), intestine, reproductive system, and glands. Such simplicity, combined to the worm’s transparency throughout its lifespan, allowed C. elegans to become the only multicellular organism with a completely established cell lineage and connectome. What’s more, these features make the worm an effective in vivo model that is amenable to whole-organism high-throughput compound screens and large-scale target validation.
Another unique advantage of C. elegans over other multicellular model organisms is that individuals can be frozen indefinitely and revived to obtain an F2 generation within a week, thus eliminating the cost of maintaining colonies of mutants.
C.elegans was the first multicellular organism to have its genome sequenced. Today, a complete and detailed C. elegans genetic map containing over 2000 loci is freely available.
Not only do the C. elegans and human genomes contain almost the same number of genes (~20,000), these two evolutionarily distant species share a surprisingly large number of cellular and molecular mechanisms. In fact, it is estimated that 65% of human disease genes have homologs in C. elegans (Baumeister R and Ge L, 2002). For diseases that can be defined on a molecular basis, C. elegans is a valuable disease model. For example, if the underlying cause of a disease is a defect in serotonergic signalling, C. elegans models can be developed to study serotonergic signalling in detail. Such models can then be pharmacologically validated and can enhance research around the serotonergic signalling network. The advantage of C. elegans over in vitro models is that instead of studying the isolated drug-receptor interaction, a functional serotonergic synapse can be studied within the context of a whole organism. The phenotypic readout of genetic and pharmacological manipulations can be measured in various ways, including the monitoring of fluorescent probes, behavioural responses such as pharyngeal pumping, and morphological changes. As a result, C. elegans has become a key model for understanding the molecular mechanisms involved in a growing number of human diseases.
The C. elegans genome is highly amenable to reverse and forward genetics, and multiple mutagenesis strategies can be used in the worm, such as whole-genome, target-selected, and gene-targeted mutagenesis (Kutscher et al, 2014). C. elegans studies are greatly facilitated by the large panel of engineering of genetic tools and genomic resources such as the RNAi-feeding library. This genetic tractability has permitted the fast generation of thousands of mutants, transgenics and knockouts, readily available at a low cost through the Caenorhabditis Genetics Center. In 2013, Thompson and colleagues published the “Million Mutations Project” in which they present a library of over 2000 mutagenized C. elegans strains, identifying mutant alleles for each of the worm’s 20,000 genes with an average of 8 non-synonymous mutations per gene and more than 16,000 insertion/deletion and copy number changes (Thompson et al., 2013).
By screening and analyzing mutants that either suppress or enhance previously characterized phenotypes, researchers are able to identify functionally interacting gene products, potentially leading to novel putative drug targets. The powerful genetics that C. elegans offers has resulted in several key scientific advances that have accelerated the biomedical research field. In 1993, the first presenilin was discovered in C. elegans. C. elegans research has further advanced the understanding of AD by identifying the presenilins as components of the -secretase complex, an important target in AD (Sundaram and Greenwald, 1993). In 1997, genetic studies in C. elegans identified negative regulators of the insulin signalling pathway (Ogg et al., 1997). One of these genes, daf-16, encodes the C. elegans orthologue of the forkhead transcription factor FOXO. Five years later, FOXO loss-of-function was found to rescue the diabetic phenotype of insulin-resistant mice (Nakae et al., 2002).
Learn more about our C. elegans products & services:
– C. elegans Transgenic Services.
– C. elegans models for disease research.
– C. elegans models for neuro disease research.
Drosophila melanogaster
The fruit fly (Drosophila melanogaster) has been instrumental to key discoveries in biomedicine. Many of the genes described as being crucial for fly development have later been shown to be important for human development as well. For example, the role played by chromosomes in heredity and the genetic control of early embryonic development was discovered using Drosophila models.
Drosophila is relatively inexpensive and easy to culture and there are very few ethical and regulatory restrictions on its use in laboratory. Like for C. elegans, the fly’s short life cycle (it takes approximately 12 days at 25°C to obtain a fertile adult fruit fly) and large brood size (120 eggs per adult and per day (Shapiro et al., 1932)) represent one of the many advantages of this model organism in biomedical research (Table 1). These advantages make Drosophila a powerful alternative to costly and time-consuming rodent experiments for unveiling the basic mechanisms involved in human diseases (Beckingham et al., 2005).
Unlike C. elegans, it is not yet possible to freeze down Drosophila gametes or embryos, thus fly strains must be carefully maintained as living stocks that need to be regularly turned over onto fresh food. In addition, female flies used in controlled genetic crosses must be “virgins”, therefore, Drosophila must be collected 8-12 hours after the culture has been cleared of adults. However, the timing of this window appears to be genotype dependent, and thus should be determined empirically for a given strain.
The adult fly has tissues and organs that are functionally equivalent to the mammalian heart, lung, kidney, gut, and reproductive tract. Drosophila’s differentiated organs, in parallel with a relative architectural simplicity, makes it one of the most powerful models to study complex mechanisms such as organogenesis and mating behaviors.
The Drosophila genome was fully sequenced in 2000. In 2001, Reiter and colleagues found that 77% of 929 genes entries known to be involved in human diseases have a recognizable match within the fruit fly genome (Reiter et al., 2001), thus further establishing Drosophila as a valuable model to study mechanisms underlying human diseases.
In the last decades, several fly models have been developed to gain insight into the mechanisms of complex diseases (Pandey and Nichols, 2011, Piazza et al., 2011, Men TT et al., 2016; Merzetti et al, 2016; Zwarts L et al., 2015 for review). For example, flies that express proteins involved in Parkinson’s, Alzheimer’s, and polyglutamine diseases such as Huntington’s chorea show cellular and behavioral phenotypes reminiscent of the symptoms of human patients (Bonini and Fortini, 2003; Iijima et al., 2004; Iijima and Iijima-Ando, 2008).
The ease and cost effectiveness to generate transgenic flies make it easy to rapidly generate models of human diseases through mutation, genetic inactivation, or misexpression of fly or human disease-causing genes and proteins.
Therefore, current and potential uses of Drosophila are expected to play a crucial role in the development of therapeutic trials aiming to test the effects of novel drugs on the evolutionarily conserved biochemical pathways that control key cellular activities.
Danio rerio
The Zebrafish (Danio rerio) is a small freshwater teleost mostly used as a model for neurodevelopmental research. Zebrafish embryos are transparent and develop externally from the mothers, offering direct access for manipulation and observation of organogenesis. Early-stage zebrafish embryos have a transparent body, making it relatively easy to collect numerous data points using high-quality imaging. For these reasons, zebrafish embryos have been proposed as an in vitro animal model for human diseases such as fetal alcohol syndrome and tuberculosis.
One limitation of zebrafish as a model organism to study human diseases is that comparatively few mutant strains are available. Zebrafish genome contains numerous duplicate genes (Milan DJ et al, 2003) and this feature often hinders the generation of knockout strains, as approaches that disrupt one gene copy likely will not disrupt the second copy. However, this limitation can now be circumvented using programmable nucleases such as (transcription activator-like effector nucleases (TALENs) and CRISPR-associated Cas9 endonucleases (Patanayak et al., 2014; Hoshijima et al, 2016). These new reverse genetic tools and the ease of transgenesis and transplantation in zebrafish contribute to more broadly investigate gene function at a systems level in zebrafish.
The zebrafish genome has been fully sequenced and its annotation is an ongoing project. A recent study indicated that 84% of the genes known to be associated with human disease have a counterpart in the zebrafish genome (Howe et al., 2013). However, the primary advantages of zebrafish (transparent embryos that develop outside of the mother) are developmentally constrained, limiting the applicability of zebrafish to study diseases that arise in adulthood.
Learn more about our Zebrafish Genome-Editing Services.
Table 2: Nobel Prizes awarded for discoveries made using Drosophila, C. elegans and zebrafish.
Conclusion
Non-mammalian model organisms are typically used in early research to deliver fast answers to a discovery problem, such as the function of a gene, or to define novel therapeutic entry points. The most popular model organisms in biological and biomedical research are the fruit fly Drosophila melanogaster, the zebrafish and the nematode C. elegans. These organisms present the advantage of being highly prolific reproducers with relatively short generation time. They are easily grown in laboratory setting and by far less expensive than murine models and more powerful than cell culture systems. In addition, thanks to the vast number of molecular and genetic tools available to study Drosophila, C.elegans and zebrafish, coupled with similarities in development and behavior, these organisms serve as powerful alternatives to mammalian model to study human genetics and diseases.
Small model systems permit systematic deciphering of the gene-gene and gene-environment interactions that influence human disorders. Through developing new and more specialized assays, small model organisms will continue to contribute new insights to the mechanisms underlying complex human diseases.
Take advantage of the CRISPR Cas9 knockout technique to delve deeper into the mechanisms underlying complex human diseases. Join us now in advancing the frontiers of scientific knowledge.
Bibliography
- Russell, W. M. S. and Burch, R. L. The Principles of Humane Experimental Technique. London: Methuen and Co. Ltd. (1959).
- Muschiol, D., Schroeder, F. & Traunspurger, W. Life cycle and population growth rate of Caenorhabditis elegans studied by a new method. BMC Ecol. 9, 14 (2009).
- Baumeister, R., Ge, L.,The worm in us – Caenorhabditis elegans as a model of human disease. Trends Biotechnol. 20, 147-148 (2002).
- Kutscher, L. M. & Shaham, S. Forward and reverse mutagenesis in C. elegans Wormbook.1.167 (2014).
- Thompson, O. et al. The million mutation project: A new approach to genetics in Caenorhabditis elegans. Genome Res. 23, 1749-1762 (2013).
- Sundaram, M. & Greenwald, I. Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics 135, 765-783 (1993).
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A., et al. The Fork head transcription factor DAF-16 transduces insulin- like metabolic and longevity signals in C. elegans. Nature 389, 994-999 (1997).
- Nakae J, Biggs WH, Kitamura T, Cavenee WK, Wright CV, Arden KC, Accili D. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. Oct;32(2):245-53 (2002).
- Shapiro, H. The rate of oviposition in the fruit fly, Drosophila. Biological Bulletin, Marine Biological Laboratory Vol. 63, No. 3, pp. 456-471 (1932).
- Beckingham KM, Armstrong JD, Texada MJ, Munjaal R, Baker DA. Drosophila melanogaster: The model organism of choice for the complex biology of multi-cellular organisms. Gravit Space Biol Bull 18: 17-29 (2005).
- Reiter, L. T., Potocki, L., Chien, S., Gribskov, M. & Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11, 1114-25 (2001).
- Udai Bhan Pandey and Charles D. Nichols. Human Disease Models in Drosophila melanogaster and the Role of the Fly in Therapeutic Drug Discovery. Pharmacol. Rev. 63, 411-436 (2011).
- Piazza, N. & Wessells, R. J. Drosophila models of cardiac disease. Prog. Mol. Biol. Transl. Sci. 100, 155-210 (2011).
- A Drosophila Model for Screening Antiobesity Agents.Men TT, Thanh DN, Yamaguchi M, Suzuki T, Hattori G, Arii M, Huy NT, Kamei K. Biomed Res Int. 2016:6293163 (2016).
- Merzetti, E. M. & Staveley, B. E. Altered expression of CG5961, a putative Drosophila melanogaster homologue of FBXO9, provides a new model of Parkinson disease. Genet. Mol. Res. 15, (2016).
- Zwarts, L. et al. The genetic basis of natural variation in mushroom body size in Drosophila melanogaster. Nat. Commun. 6, 10115 (2015).
- Bonini, N. M., Fortini, M. E. Human neurodegenerative disease modeling using Drosophila. Annu. Rev. Phytopathol. 40, 381-410 (2002).
- Iijima, K. et al. Dissecting the pathological effects of human A beta 40 and A beta 42 in Drosophila: A potential model for Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 101, 6623-6628 (2004).
- Iijima, K. & Iijima-Ando, K. Drosophila models of Alzheimer’s amyloidosis: the challenge of dissecting the complex mechanisms of toxicity of amyloid-beta 42. J. Alzheimers. Dis. 15, 523-540 (2008).
- Milan, D. J., Peterson, T. A., Ruskin, J. N., Peterson, R. T. & MacRae, C. A. Drugs that induce repolarization abnormalities cause bradycardia in zebrafish. Circulation 107, 1355-1358 (2003).
- Pattanayak, V., Guilinger, J. P. & Liu, D. R. Determining the specificities of TALENs, Cas9, and other genome-editing enzymes. Methods Enzymol. 546, 47-78 (2014).
- Hoshijima, K. et al. Precise Editing of the zebrafish genome made simple and efficient. Dev. Cell 36, 654-667 (2016).
- Howe, K. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498-503 (2013).
Related Questions
Which alternate methods are now available instead of traditional use of mice and monkey models for biomedical research?
Due to many ethical and budgetary concerns about traditional methods, new strategies and systems have been adapted. Some of these models are cell culture, Danio rerio (zebrafish), Drosophila melanogaster (fruit fly) and Caenorhabditis elegans (worm).
How do I choose the research model for my study?
The selection of which organisms to use as a research model depends on the nature of the disease or mechanism being studied and the scientific questions being asked. Please review the table 1 in the article.
What is the limitation of using cell culture for my research?
Although cell culture models represent a fast and powerful tool for proof of concept, results obtained in vitro often poorly correlate with in vivo mechanisms. This is due in part to the fact the mechanisms studied in vitro occur in a simplified, highly controlled but often poorly understood intracellular and extracellular environment with little physiological relevance. Moreover, the use of dissociated cells precludes access to the behavioral output of biological mechanisms.
Which is the medium of growth for C. elegans and what diet should be maintained?
C. elegans can be grown on agar medium in Petri dishes, where it can be maintained on a simple diet of Escherichia coli bacteria, which makes this model very easy and cost-effective.
What are some of the benefits of using C. elegans model for my research?
Besides being cost-effective, C. elegens have a short life cycle for over two weeks, which is very useful for studying their development. C. elegans has been used widely for answering questions related to neurodegenerative diseases. It also provides models for many human diseases including congenital heart disease and kidney disease. Another unique advantage of C. elegans over other multicellular model organisms is that individuals can be frozen indefinitely and revived to obtain an F2 generation within a week, thus eliminating the cost of maintaining colonies of mutants.
What makes C. elegans an excellent system to study neurological disorders?
The nervous system of C. elegans is very simple and uses most of the neurotransmitters presents in the mammalian brain. These characteristics make it a desirable model for studying the neuronal connectome and cellular mechanisms involved in complex disorders.
What are the advantages of using Drosophila melanogaster model for my studies?
Drosophila is relatively inexpensive and easy to culture and there are very few ethical and regulatory restrictions on its use in laboratory. Like for C. elegans, the fly’s short life cycle (it takes approximately 12 days at 25°C to obtain a fertile adult fruit fly) and large brood size (120 eggs per adult and per day represent one of the many advantages of this model organism in biomedical research. These advantages make Drosophila a powerful alternative to costly and time-consuming rodent experiments for unveiling the basic mechanisms involved in human diseases.
Why are zebrafish useful as models of development?
Zebrafish model has been used for decades to study development and pathophysiology. Zebrafish share a largely conserved physiology and anatomy with mammals. A recent study indicated that 84% of the genes known to be associated with human disease have a counterpart in the zebrafish genome. Furthermore, their genome sizes are approximately 20-40% of the mammalian genome, and it makes them the only vertebrates available for large-scale mutagenesis.
Which countries have comparatively larger population of Zebrafish in their freshwaters?
Zebrafish is a small tropical freshwater fish present in river basins of India, Northern Pakistan, Nepal, Bhutan, and South Asia.