In recent years, the nematode C. elegans has emerged as a model for systematic dissection of the molecular basis of tumorigenesis. Many cancer genes and pathways are highly conserved and often easier to parse in C. elegans as the gene families involved contain fewer members, reducing genetic redundancy. This case study illustrates how C. elegans were used to decipher breast cancer.
Breast cancer is the most common malignancy in women of the Western world, affecting up to 10% of the female population.1 BRCA1 and BRCA2 are human genes that produce tumor suppressor proteins. Together, BRCA1 and BRCA2 mutations account for about 20-25% of hereditary breast cancers, about 5-10% percent of all breast cancers and around 15% of ovarian cancers. Mutations in BRCA1 are associated with increased susceptibility to breast and ovarian cancers.2
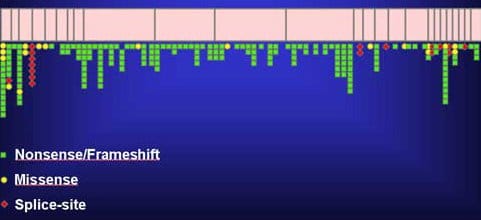 Figure 1: Representation of the various mutations on the BRCA1 gene associated with cancer (National Cancer Institute).
Figure 1: Representation of the various mutations on the BRCA1 gene associated with cancer (National Cancer Institute).
Like in human cells, in C. elegans germline, apoptosis can be activated in response to pathogenic microorganism invasion or DNA-damaging agents and genotoxic stress. In 2006, Polanowska et al. showed that deletion of brc-1, the C. elegans orthologue of tumor suppressor BRCA-1, share many of the functional hallmarks of BRCA1-deficient human cells, such as specificity for the UbcH5c homolog and recruitment to sites of DNA damage.3 Most BRCA1 homozygous mutations caused embryonic lethality in mice, hindering in vivo analysis of tumor suppressor functions of this tumor suppressor.
One great advantage of C. elegans as a breast cancer model is that worms bearing a homozygous deletion of brc-1 are viable, thus permitting extensive reverse genetic analysis.
In order to identify genes that could be targeted in human patients to selectively kill tumors defective in the BRCA pathway, researchers performed a genome-wide screen by systematically inactivate all 19,500 C. elegans genes. They identified 20 genes whose depletion confers synthetic lethality in combination with brc-1.
Among those genes, was pme-2, the C. elegans counterparts of PARP, whose inhibition selectively kills BRCA defective tumor cells in human patients.4 Three years later, Kato et al. showed that the mir-34 microRNA is required for the DNA damage response both in vivo in C. elegans and in vitro in human breast cancer cells.5 It is interesting to notice that miR-34, the founding member of an evolutionarily conserved miRNA family found in diverse species, including humans, was first identified in C. elegans.
Human breast/ovarian cancers have also been linked to X chromosome abnormalities.6 In particular, most breast cancers lack markers of a normal inactive X chromosome and correlate with a duplication of the active X chromosome. Interestingly, male C. elegans are determined by a single X chromosome, while hermaphrodites are determined by two copies. Therefore, X chromosome nondisjunction leads to higher frequencies of males.7
It is reasonable to assume that the human orthologs of genes associated with high frequency of males are good candidate genes for involvement in breast/ovarian cancers. In particular, studies have shown that some of these genes, such as GCC2, PIGA, WDHD1 and SEH1L are implicated as breast cancer candidate genes.
See C. elegans genes used as experimental models for investigating other human diseases.
- Alberg AJ, Helzlsouer KJ. Epidemiology, prevention, and early detection of breast cancer. Curr Opin Oncol 9: 505–511 (1997).
- Silver D. P., Livingston D. M. Mechanisms of BRCA1 tumor suppression. Cancer Discov. 2: 679–684 (2012).
- Polanowska, J., Martin, J. S., Garcia-Muse, T., Petalcorin, M. I. R. & Boulton, S. J. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 25, 2178–88 (2006).
- Petalcorin, M., Martin, J. & Boulton, S. Genome-wide RNAi to identify genes that confer synthetic lethality with BRCA1. Breast Cancer Res. 8, S10 (2006).
- Kato, M. et al. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28, 2419–24 (2009).
- Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9:121–132 (2006).
- Hodgkin, J., Horvitz, H. R., & Brenner, S. . Nondisjunction Mutants of the Nematode Caenorhaditis elegans. Genetics, 91(1), 67–94 (1979).


